Research Article
Insilico investigation of TNFSF10 signaling cascade in ovarian serous cystadenocarcinoma

Asima Tayyeb1* and Zafar Abbas Shah2
1ASchool of Biological Sciences, University of the Punjab, Lahore, Pakistan2Department of Bioinformatics, Hazara University, Mansehra, Pakistan
*Address for Correspondence: Dr. Asima Tayyeb, School of Biological Sciences, Quaid-e-Azam Campus, University of the Punjab, Lahore, Pakistan, Email: [email protected]; [email protected]
Dates: Submitted: 22 May 2019; Approved: 01 July 2019; Published: 02 July 2019
How to cite this article: Tayyeb A, Shah ZA. Insilico investigation of TNFSF10 signaling cascade in ovarian serous cystadenocarcinoma. Arch Cancer Sci Ther. 2019; 3: 025-034. DOI: 10.29328/journal.acst.1001005
Copyright License: © 2019 Tayyeb A, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Ovarian cancer; TNFSF10; TNFSF10; FAS
Abstract
The ovarian serous Cystadenocarcinoma shared large number of deaths in gynecologic carcinoma. It has various numbers of molecular events from initiation to progression and at advance stage, surgery is the end product of such molecular signaling. We assess in this study the whole mechanistic view of TNFSF10 network which has the ideal apoptotic causing identity. We used fresh insilico strategy to uncover the secrets and inter-links from its protein-protein interaction complex. We retrieved the TNFSF10 signaling network from STRING database (www.string-db.org). The network contains 25 nodes and 152 edges with clustering presentation. After retrieval, we performed gene enrichment and characterization analysis of network from WebGestalt toolkit (www.webgestalt.com). Finally, we examined the participation of whole network in ovarian cancer progression from cBioPortal, a cancer genomic data portal (www.cbioportal.org). Our results showed that majority of cases have loss of function of death receptors (DR4 and DR5) that are the main unit of initiation of apoptotic signaling. Most of downstream signaling members showed amplification that regulates cell proliferative pathways including NFkB pathway. TNFSF10 cluster has loss of function and in future it gain attention for further research studies to discover its interactome level view for valuable therapy. FAS cluster has large number of members and majority showed amplification rendering them as co-targets for combinational drug designing.
Introduction
Tumor Necrosis Factor Super Family 10 (TNFSF10) is a member of TNF super family positioned on chromosome 3 that code protein of 20kDa which consist of 4 intron and 5 exon. TNFSF10 protein has 281 amino acids regulated mainly as a transmembrane protein and releases protein as a soluble form after C-terminus cleavage [1-3]. The primary function of TNFSF10/TRAIL is the induction of apoptotic process in transformed cells and mostly functional in immune cells [4-6]. TNFSF10 trigger the extrinsic apoptotic mechanisms by binding with death receptor 4 (DR4) and death receptor 5 (DR5). It also regulates NFkB and MAPK8 pathways that play key role in cell survival and growth [7]. TNFSF10 perform vital role in development of death-inducing signaling complex (DISC) that contains CFLAR, FADD, CASP8 and death receptors [8]. In carcinogenesis metastatic cells block the function of TNFSF10 of DISC development by inhibition of decoy receptors including DCR1, DCR2, OPG (Osteoprotegerin) and overexpression of anti-apoptotic agents like BIRC2, CFLAR and NFkB [9-11]. Some studies explore TGM2, EGFR, MCL1 and PTGS2 pathways that counteract TNFSF10 activity [12,13].
To study the fundamental significance of TNFSF10/TRAIL in progression of ovarian carcinoma we practice novel insilico approach to determine not only its individual expression but also with network level holistic view. In this approach we uncovered pathway interactors to find out therapeutic targets.
Insilico protocol
We retrieved TNFSF10 signaling cascade from STRING database (www.string-db.org) which is huge collection of protein-protein interaction networks with different topological, ontological and statistical tools that perform function on scoring pattern which showed the authenticity of a network. We process the enrichment and characterization analysis of a TNFSF10 cascade in WebGestalt toolkit (www.webgestalt.com) that gives us its ontological view with multiple characteristics for its deep understanding. We determined the cascade participation in disease progression by WebGestalt. Finally we use cBioPortal a cancer genome platform (www.cbioportal.org) that contains large number of cancer datasets with fine graphical presentation, enrichment tools, topological analysis tools and mutation identifiers that comprehensively find out oncogenic activities.
Results
TNFSF10 Signaling cascade mining from STRING database
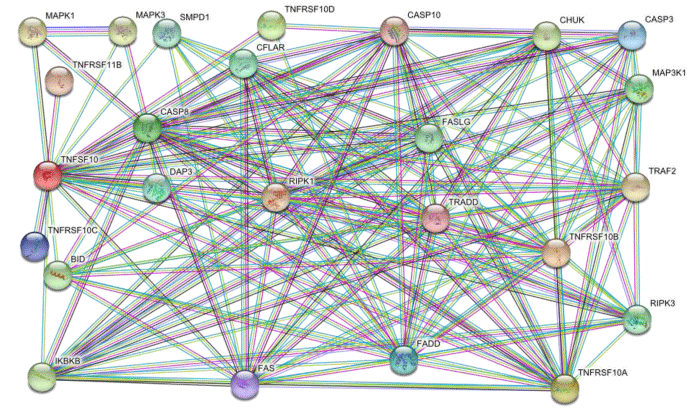
TNFSF10 signaling cascade retrieved from STRING database. The signaling cascade contain various topological characters such as 25 nodes, 152 edges, 12.2 average node degree and 0.879 average local clustering coefficient. The signaling cascade showed the properties of scalable vector network. The cascade has strong protein-protein interactions of highest confidence score of 0.900. The hub proteins are densely connected with other proteins in cascade (Figure 1.1).
Figure 1.1: TNFSF10 signaling cascade of 25 nodes and 152 edges which showed proteins and their interactions respectively. The multiple color lines indicate various interaction types among proteins on system level.
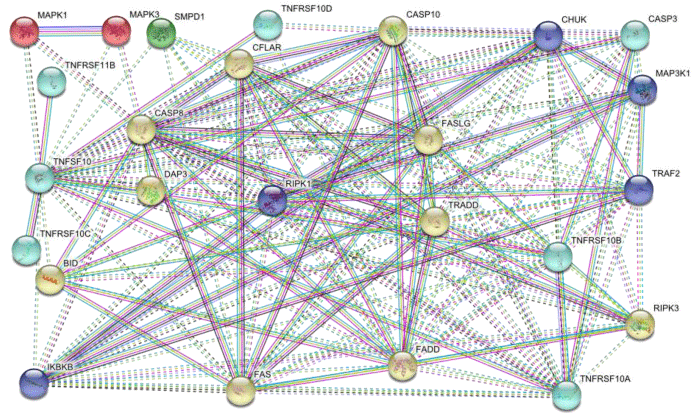
STRING database allowed us to draw functionally associated clusters that bind with each other in the form of inter cluster association and inside cluster association is intra cluster that showed the strength of interaction. The clustering of a network has enormous significance for drug targeting. We obtain 5 clusters by K-MEANS clustering algorithm of STRING database. First TNFSF10 cluster contain seven members including TNFRSF11B, TNFRSF10C, TNFRSF10A, TNFRSF10B, CASP3 and TNFRSF10D. Second IKBKB cluster contains five members including RIPK1, TRAF2, CHUK and MAP3K1. Third MAPK1 cluster has MAPK3. Forth SMPD1 cluster consist on only one member. Fifth CASP8 cluster is the largest cluster that contains ten members including FAS, BID, DAP3, CFLAR, FASLG, TRADD, FADD, RIPK3 and CASP10 (Figure 1.2).
Figure 1.2: TNFSF10 cascade clusters in which dotted lines showed inter cluster and straight lines indicate intra cluster association.
TNFSF10 signaling cascade components gene enrichment ontology analysis by WebGestalt toolkit
TNFSF10 signaling cascade components involved in most important programmed cell death process that help in organogenesis at embryonic level and encounter tumorigenesis. In apoptosis cell undergo certain steps that lead to its death in which cell stress causing autophagy is known as intrinsic pathway and due to cellular signaling cell death is called extrinsic pathway (Table 1.1).
| Table 1.1: The WebGestalt process of biological characterization analysis of TNFSF10 pathway. | ||
| S. NO | BIOLOGICAL PROCESSES | NO. OF PROTEINS |
| 1 | Extrinsic apoptotic signaling pathway | 13 |
| 2 | Apoptotic process | 23 |
| 3 | Programmed cell death | 23 |
| 4 | Death | 23 |
| 5 | Cell death | 23 |
| 6 | Apoptotic signaling pathway | 13 |
| 7 | Regulation of execution phase of apoptosis | 13 |
| 8 | Activation of cysteine-type endopeptidase activity involved in apoptotic process | 11 |
| 9 | Activation of cysteine-type endopeptidase activity | 11 |
TNFSF10 pathway components are involved in various molecular mechanisms to initiate apoptosis. They interact with death receptors, decoy receptors, Osteoprotegerin and development of DISC by web like binding with FADD, CASP8 and other pro-apoptotic ingredients (Table 1.2).
| Table 1.2: WebGestalt process of molecular function enrichment analysis of TNFSF10 cascade. | ||
| S. NO | MOLECULAR FUNCTIONS | NO. OF PROTEINS |
| 1 | Tumor necrosis factor receptor superfamily binding | 7 |
| 2 | TRAIL binding | 4 |
| 3 | Tumor necrosis factor receptor binding | 5 |
| 4 | Cytokine receptor binding | 7 |
| 5 | Protein binding | 24 |
| 6 | Enzyme binding | 11 |
| 7 | Death receptor binding | 3 |
| 8 | Receptor signaling protein serine/threonine kinase activity | 4 |
| 9 | Protein serine/threonine kinase activity | 7 |
| 10 | Cysteine-type endopeptidase activity | 4 |
TNFSF10 pathway components are widely dispersed in cellular microenvironment from cell membrane to cytosol to mitochondria that trigger apoptotic signaling pathways that ultimately affect on gene regulation in nucleus (Table 1.3).
| Table 1.3: WebGestalt process of cell localization ontology analysis of TNFSF10 cascade members. | ||
| S. NO | CELL LOCALIZATION | NO. OF PROTEINS |
| 1 | Death-inducing signaling complex | 5 |
| 2 | Membrane raft | 9 |
| 3 | CD95 death-inducing signaling complex | 3 |
| 4 | Cytosol | 15 |
| 5 | CD40 receptor complex | 3 |
| 6 | Plasma membrane part | 13 |
| 7 | Receptor complex | 5 |
| 8 | Plasma membrane | 17 |
| 9 | Cell periphery | 17 |
| 10 | I kappaB kinase complex | 2 |
TNFSF10 components are involved in various other intra cellular signaling pathways that are the regulators of cell proliferation, migration, differentiation, survival, growth, death, stress response and metastasis (Table 1.4).
| Table 1.4: WebGestalt process of characterization of pathway to pathway interaction analysis of TNFSF10 cascade. | ||
| S. NO | KEGG PATHWAYS | NO. OF PROTEINS |
| 1 | Apoptosis | 18 |
| 2 | Natural killer cell mediated cytotoxicity | 11 |
| 3 | RIG-I-like receptor signaling pathway | 9 |
| 4 | Chagas disease | 9 |
| 5 | Pathways in cancer | 11 |
| 6 | MAPK signaling pathway | 9 |
| 7 | Toll-like receptor signaling pathway | 7 |
| 8 | Hepatitis C | 7 |
| 9 | Cytokine-Cytokine receptor interaction | 8 |
| 10 | Alzheimer disease | 7 |
This is most appropriate result to understand the significance of TNFSF10 signaling components in disease causing and progression. This signaling network acts as a protective unit but due to its deregulation several viral and pathogenic problems are developed. The abnormal signaling observed in certain carcinomas and immune diseases (Table 1.5).
| Table 1.5: WebGestalt process to characterize TNFSF10 complex components in disease. | ||
| S. NO | Disease enrichment analysis | NO. OF PROTEINS |
| 1 | Necrosis | 23 |
| 2 | Death | 21 |
| 3 | Lymphoproliferative disorder s | 12 |
| 4 | Cancer or viral infections | 14 |
| 5 | Drug interaction with drug | 11 |
| 6 | Brain death | 8 |
| 7 | Lymphatic diseases | 10 |
| 8 | Lentivirus infections | 10 |
| 9 | Hodgkin disease | 8 |
| 10 | Infection | 10 |
TNFSF10 signaling cascade core analysis of exploring its involvement in carcinoma by cBioPortal
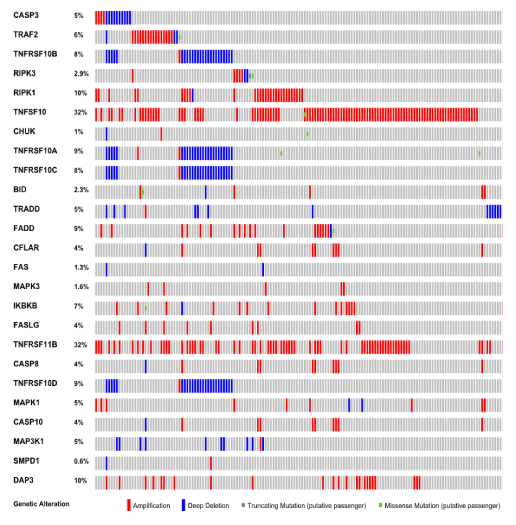
We use cBioPortal a cancer genome platform that provide large collection of cancer dataset with attractive graphical presentation and comprehensive oncogene detection with accurate ratio and site of mutation. We gain a dataset of ovarian serous Cystadenocarcinoma (TCGA, Provisional) that contain 311 samples in which 214 cases showed alteration in which 182 cases showed amplification, 20 cases showed deletion, 10 cases showed multiple alteration and 2 cases indicate mutation. The whole signaling cascade showed 68.8% alteration in which 58.5% amplification, 6.4% deletion, 3.2% multiple alteration and 0.6% mutation (Figure 1.3).
Discussion
The process of eradicating the harmful cells through internal/external stimuli is called apoptosis that undergo two basic signaling pathways which are intrinsic and extrinsic pathways. In mitochondria Bcl2 proteins family trigger and regulate intrinsic pathway while TNF family with receptors trigger and regulate extrinsic apoptotic pathway [14-17]. Due to sequence similarity of TNF and FASLG extracellular domain first time TNFSF10 was explored that has four receptors of his own family that are TNFRSF10A/DR4, TNFRSF10B/DR5, TNFRSF10C/DCR1 and TNFRSF10D/DCR2. Both death receptors triggered apoptosis and decoy receptors inhibit TNFSF10 induced apoptosis [18]. TNSFSF10 interact with its death receptors on cell membrane that deploy FADD and CASP8 that developed DISC (death-inducing signaling complex). The deployment and activation of CASP8 trigger the regulation of CASP3 that initiate comprehensive apoptotic process [19,20]. The activated CASP8 also target the BID that undergo firstly in cleavage and then stimulate the BAX and BAK pro-apoptotic components that are releasing cause of Cytochrome C. The apoptosome development occurred by the combination of ATP, CASP9, Cytochrome C and APAF1 that trigger the cleavage of CASP3 that amplify the whole TNFSF10 induced apoptotic signaling [21-27]. Several studies reported the contribution of TNFSF10 mediated signaling cascade in meningitis, inflammation, diabetes, asthma, immune diseases and carcinogenesis [28-35]. In this work we apply fresh insilico procedure that determines the holistic view of TNFSF10 mediated apoptotic signaling complex. We interestingly identified TNFSF10 32% overexpression that is the possible effect of immune microenvironment interaction with ovarian tumor cells. This expression indicates the better survival of ovarian cancers which is the confirmation of earlier studies [36]. We determine that TNFSF10 death receptors showed loss of function in context of deletion that are located on chromosome 8 which undergo mostly loss of function due to deletion or mutation. They contain highly conserved region of 95 amino acids of death domains which interact with TNFSF10 and initiate apoptotic process. Their loss of function is reported in several carcinomas including head and neck, breast and lung carcinoma [37-42]. The deletion of death receptors finishes the ability of DISC development that is the organizer of apoptotic signaling [43]. Both receptors are functionally associated with each other and loss of function makes TNFSF10 functionally insignificant [44,45]. In ovarian serous carcinoma these are the key drug targets for normal development of DISC and tumor cells eradication. The decoy receptors showed deletion that normally heterotrimeric complex that prevent apoptosis via deactivation of CASP8 [46-49]. TNFRSF11B showed 32% overexpression which is secreted protein with lack of transmembrane domain and participate in inhibition of TNFSF10 mediated pathway [50]. More importantly CASP3 showed deletion in most ovarian serous carcinoma cases that is the end target of activated CASP8 for proper initiation of apoptotic signaling. TNFSF10 cluster contains mostly those members that undergo loss of function due to deletion so required further research to explore their network view that are the causing agent of deletion.
Our work determined the IKBKB overexpression that is the regulator of NFkB signaling pathway and required for IKBα/β phosphorylation [51]. The IKBKB cluster also has the overexpression of RIPK1 and TRAF2 that are the inducer of NFkB anti-apoptotic proliferative pathway [52,53]. This cluster explains the role of cell proliferation by NFkB signaling through amplified TNFSF10 expression. In this study the FAS cluster has large number of members that showed its functional diversity. FAS showed loss of function in context of deletion, it played a vital role in apoptotic signaling mechanisms of immune cells, destruction of cells by T-Cell-mediated pathway and its deregulation reported in various diseases [54,55]. CFLAR and FASLG both is negative regulator of TNFSF10 pathway that showed amplification in results. TRADD showed loss of function which is the main linker of FADD and downstream signaling activators of apoptosis [56]. FADD and CASP8 both showed amplification but due to loss of function of various TNFSF10 cascade key members they behave functionally inactive in pro-apoptotic context and prevent TNFR1 signaling [57-58]. The MAPK cluster showed overexpression that involved in cell survival and proliferation in response of various forms of stress.
Our study confirmed the dual and unique role of TNFSF10 mediated signaling in which they induce cell proliferation signals by overexpression of FADD, RIP1, TRAF2, IKBKB and CFLAR which ultimately activate the NFkB signaling pathway which further regulate ErK2, JNK, MAPK and PI3K signaling proliferative pathways. Several cancer studies reported such role in progression and promotion of metastasis [59-65]. In this approach we determined the TNFSF10 mediated signaling cascade participation in ovarian serous carcinoma and find interesting links of anti-apoptotic /proliferative channels. This effort addresses the basic theme of TNFSF10 signaling pathway and increase the comprehension of its patho-physiology before their therapy.
Conclusion
System biology deals the phenomenon based on inter-link events in which signaling cascades perform a major role in regulation of system level mechanisms. Day by day secretes of interactomics are uncovered by insilico studies. In this study we examined TNFSF10 signaling network to assess its core mechanistic view for its functioning in progression of carcinogenesis. Interestingly we identified several unique interactions that showed overexpression and promote cell proliferation which is the negative sign of this pathway. There is complex story of TNFSF10 network members that show diverse sensitivity to stress and mutagens. We analyzed the network on cluster based in which TNFSF10 cluster show maximum loss of function that is main cause of promotion of tumorigenesis and majority of members of all other clusters of network showed overexpression that promote cell survival and regulation of proliferative pathways. In future our study proved as a repository of markers for TNFSF10 network based drug designing.
References
- Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, et al. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996; 271:12687-12690. Ref.: http://bit.ly/2Nso1Su
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995; 3: 673-682. Ref.: http://bit.ly/2LxPDmv
- Kimberley FC, Screaton GR. Following a TRAIL: update on a ligand and its five receptors. Cell Res. 2004; 14: 359. Ref.: http://bit.ly/327eofe
- Diehl GE, Yue HH, Hsieh K, Kuang AA, Ho M, et al. TRAIL-R as a negative regulator of innate immune cell responses. Immunity. 2004; 21: 877-889. Ref.: http://bit.ly/2LulNzj
- Cretney E, Takeda K, Yagita H, Glaccum M, Peschon JJ, et al. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J Immunol. 2002; 168: 1356-1361. Ref.: http://bit.ly/2xnjPZf
- Abdulghani J, El-Deiry WS. TRAIL receptor signaling and therapeutics. Expert Opin Ther Targets. 2010; 14: 1091-1108. Ref.: http://bit.ly/2JpayFw
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001; 15: 2922-2933. Ref.: http://bit.ly/2FM402U
- Lin Y, Devin A, Cook A, Keane MM, Kelliher M, et al. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IκB kinase and c-Jun N-terminal kinase. Mol Cell Biol. 2000; 20: 6638-6645. Ref.: http://bit.ly/2XFqZH3
- Wajant H. TRAIL and NFκB signaling—a complex relationship. Vitamins & Hormones. 2004; 67: 101-132. Ref.: http://bit.ly/2YptNoF
- Wajant H, Gerspach J, Pfizenmaier K. Tumor therapeutics by design: targeting and activation of death receptors. Cytokine & growth factor reviews. 2005; 16: 55-76. Ref.: http://bit.ly/2ROkbS2
- Wang X, Chen W, Zeng W, Bai L, Tesfaigzi Y, et al. Akt-mediated eminent expression of c-FLIP and Mcl-1 confers acquired resistance to TRAIL-induced cytotoxicity to lung cancer cells. Mol Cancer Ther. 2008; 7: 1156-1163. Ref.: http://bit.ly/304lz5Y
- Chen W, Bai L, Wang X, Xu S, Belinsky SA, et al. Acquired activation of the Akt/cyclooxygenase-2/Mcl-1 pathway renders lung cancer cells resistant to apoptosis. Mol Pharmacol. 2010; 77: 416-423. Ref.: http://bit.ly/2LtjZql
- Li Z, Xu X, Bai L, Chen W, Lin Y. Epidermal growth factor receptor-mediated tissue transglutaminase overexpression couples acquired tumor necrosis factor-related apoptosis-inducing ligand resistance and migration through c-FLIP and MMP-9 proteins in lung cancer cells. J Biol Chem. 2011; 286: 21164-21172. Ref.: http://bit.ly/2XhrX84
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004; 116: 205-219. Ref.: http://bit.ly/323JoMW
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000; 100: 57-70. Ref.: http://bit.ly/2NskGCE
- Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002; 108: 153-164. Ref.: http://bit.ly/2ROrOYM
- Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005; 5: 876. Ref.: http://bit.ly/326G83F
- Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002; 2: 420-430. Ref.: http://bit.ly/2XK6icY
- Falschlehner, C., Emmerich, C.H., Gerlach, B. and Walczak, H., 2007. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol. 2007; 39: 1462-1475. Ref.: http://bit.ly/2YqbYpC
- Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999; 11: 255-260. http://bit.ly/2FNcIxE
- Keogh SA, Walczak H, Bouchier-Hayes L, Martin SJ. Failure of Bcl‐2 to block cytochrome c redistribution during TRAIL‐induced apoptosis. FEBS letters. 2000; 471: 93-98. Ref.: http://bit.ly/2RQt8Kt
- Walczak H, Bouchon A, Stahl H, Krammer PH. Tumor necrosis factor-related apoptosis-inducing ligand retains its apoptosis-inducing capacity on Bcl-2-or Bcl-xL-overexpressing chemotherapy-resistant tumor cells. Cancer Res. 2000; 60: 3051-3057. Ref.: http://bit.ly/2FOEr10
- Belka C, Schmid B, Marini P, Durand E, Rudner J, et al. Sensitization of resistant lymphoma cells to irradiation-induced apoptosis by the death ligand TRAIL. Oncogene. 2001; 20: 2190-2196. Ref.: http://bit.ly/2RNyFlf
- Hinz S, Trauzold A, Boenicke L, Sandberg C, Beckmann S, et al. Bcl-XL protects pancreatic adenocarcinoma cells against CD95-and TRAIL-receptor-mediated apoptosis. Oncogene. 2000; 19: 5477-5486. Ref.: http://bit.ly/2XlUvT0
- Fulda S, Meyer E, Debatin KM. Inhibition of TRAIL-induced apoptosis by Bcl-2 overexpression. Oncogene. 2002; 21: 2283. Ref.: http://bit.ly/2RNzfiV
- Zhang XD, Borrow JM, Zhang XY, Nguyen T, Hersey P. Activation of ERK1/2 protects melanoma cells from TRAIL-induced apoptosis by inhibiting Smac/DIABLO release from mitochondria. Oncogene. 2003; 22: Ref.: http://bit.ly/2RKSbyA
- Rudner J, Jendrossek V, Lauber K, Daniel PT, Wesselborg S, et al. Type I and type II reactions in TRAIL-induced apoptosis-results from dose-response studies. Oncogene. 2005; 24: 130-140. Ref.: http://bit.ly/2JjEoLJ
- Secchiero P, Zauli G. Tumor necrosis factor-related apoptosis-inducing ligand and the regulation of hematopoiesis. Curr Opin Hematol. 2008; 15: 42-48. Ref.: http://bit.ly/2NsfpuT
- Janssen EM, Droin NM, Lemmens EE, Pinkoski MJ, Bensinger SJ, et al. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005; 434: 88. Ref.: http://bit.ly/2xmWsyP
- Weckmann M, Collison A, Simpson JL, Kopp MV, Wark PA, et al. Critical link between TRAIL and CCL20 for the activation of TH2 cells and the expression of allergic airway disease. Nat Med. 2007; 13: 1308. Ref.: http://bit.ly/2XjAL24
- Cretney E, Shanker A, Yagita H, Smyth MJ, Sayers TJ. TNF-related apoptosis-inducing ligand as a therapeutic agent in autoimmunity and cancer. Immunol Cell Biol. 2006; 84: 87. Ref.: http://bit.ly/2FN9kTE
- Hilliard B, Wilmen A, Seidel C, Liu TS, Göke R, et al. Roles of TNF-related apoptosis-inducing ligand in experimental autoimmune encephalomyelitis. J Immunol. 2001; 166: 1314-1319. Ref.: http://bit.ly/2xmWDdt
- Lamhamedi-Cherradi SE, Zheng S, Tisch RM, Chen YH. Critical roles of tumor necrosis factor–related apoptosis-inducing ligand in type 1 diabetes. Diabetes. 2003; 52: 2274-2278. Ref.: http://bit.ly/2LxUrIG
- Zheng SJ, Wang P, Tsabary G, Chen YH. Critical roles of TRAIL in hepatic cell death and hepatic inflammation. J Clin Invest. 2004; 113: 58-64. Ref.: http://bit.ly/2xoU7nl
- Hoffmann O, Priller J, Prozorovski T, Schulze-Topphoff U, Baeva N, et al. TRAIL limits excessive host immune responses in bacterial meningitis. J Clin Invest. 2007; 117: 2004-2013. Ref.: http://bit.ly/2KRyUuP
- Lancaster JM, Sayer R, Blanchette C, Calingaert B, Whitaker R, et al. High expression of tumor necrosis factor-related apoptosis-inducing ligand is associated with favorable ovarian cancer survival. Clin Cancer Res. 2003; 9: 762-766. Ref.: http://bit.ly/2KRyUuP
- Wright K, Wilson PJ, Kerr J, Do K, Hurst T, et al. Frequent loss of heterozygosity and three critical regions on the short arm of chromosome 8 in ovarian adenocarcinomas. Oncogene. 1998; 17:1185-1188. Ref.: http://bit.ly/2XhEb0H
- Pribill I, Speiser P, Leary J, Leodolter S, Hacker NF, et al. High frequency of allelic imbalance at regions of chromosome arm 8p in ovarian carcinoma. Cancer Genet Cytogenet. 2001; 129: 23-29. Ref.: http://bit.ly/2KRHlq3
- Pan G, Ni J, Wei YF, Yu G, Gentz R, et al. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997; 277: 815-818. Ref.: http://bit.ly/2LxWxZ4
- Lee SH, Shin MS, Kim HS, Lee HK, Park WS, et al. Alterations of the DR5/TRAIL receptor 2 gene in non-small cell lung cancers. Cancer Res. 1999; 59: 5683-5686. Ref.: http://bit.ly/2Lr6ZS2
- Lee SH, Shin MS, Kim HS, Lee HK, Park WS, et al. Somatic mutations of TRAIL-receptor 1 and TRAIL-receptor 2 genes in non-Hodgkin's lymphoma. Oncogene. 2001; 20: 399-403. Ref.: http://bit.ly/2KWycwq
- Fisher MJ, Virmani AK, Wu L, Aplenc R, Harper JC, et al. Nucleotide substitution in the ectodomain of trail receptor DR4 is associated with lung cancer and head and neck cancer. Clin Cancer Res. 2001; 7:1688-1697. Ref.: http://bit.ly/2XHWikq
- Shin MS, Kim HS, Lee SH, Park WS, Kim SY, et al. Mutations of tumor necrosis factor-related apoptosis-inducing ligand receptor 1 (TRAIL-R1) and receptor 2 (TRAIL-R2) genes in metastatic breast cancers. Cancer research 2001; 61: 4942-4946. Ref.: http://bit.ly/2XhNSft
- Bin L, Thorburn J, Thomas LR, Clark PE, Humphreys R, et al. Tumor-derived mutations in the TRAIL receptor DR5 inhibit TRAIL signaling through the DR4 receptor by competing for ligand binding. J Biol Chem. 2007; 282: 28189-28194. http://bit.ly/2Xi62CJ
- MacFarlane M, Kohlhaas SL, Sutcliffe MJ, Dyer MJ, Cohen GM. TRAIL receptor-selective mutants signal to apoptosis via TRAIL-R1 in primary lymphoid malignancies. Cancer Res. 2005; 65: 11265-11270. Ref.: http://bit.ly/305SKWF
- van der Sloot AM, Tur V, Szegezdi E, Mullally MM, Cool RH, Samali A, et al. Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci U S A. 2006; 103: 8634-8639. Ref.: http://bit.ly/324Gzvm
- Sanlioglu AD, Karacay B, Koksal IT, Griffith TS, Sanlioglu S. DcR2 (TRAIL-R4) siRNA and adenovirus delivery of TRAIL (Ad5hTRAIL) break down in vitro tumorigenic potential of prostate carcinoma cells. Cancer Gene Ther. 2007; 14: 976-984. Ref.: http://bit.ly/2No8hzI
- Sanlioglu AD, Korcum AF, Pestereli E, Erdogan G, Karaveli S, et al. TRAIL Death Receptor–4 Expression Positively Correlates With the Tumor Grade in Breast Cancer Patients With Invasive Ductal Carcinoma. Int J Radiat Oncol Biol Phys. 2007; 69: 716-723. Ref.: http://bit.ly/2RU8kC8
- Riccioni R, Pasquini L, Mariani G, Saulle E, Rossini A, et al. TRAIL decoy receptors mediate resistance of acute myeloid leukemia cells to TRAIL. Haematologica. 2005; 90: 612-624. Ref.: http://bit.ly/2Xi7yEV
- Mérino D, Lalaoui N, Morizot A, Schneider P, Solary E, et al. Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Mol Cell Biol. 2006; 26: 7046-7055. Ref.: http://bit.ly/30eCRh1
- Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem. 1998; 273: 14363-14367. Ref.: http://bit.ly/2YsMXdl
- Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, et al. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000; 12: 419-429. Ref.: http://bit.ly/2XCTJ3e
- Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, et al. Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997; 7: 715-725. Ref.: http://bit.ly/2XoEdbN
- Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996; 4:387-396. Ref.: http://bit.ly/2Xhytfg
- Nagata S. Fas ligand-induced apoptosis. Annu Rev Genet.1999; 33: 29-55. Ref.: http://bit.ly/2YsNdsP
- Peter ME, Krammer PH. The CD95 (APO-1/Fas) DISC and beyond. Cell Death Differ. 2003; 10: 26-35. Ref.: http://bit.ly/2YnfX6p
- Song JH, Tse MC, Bellail A, Phuphanich S, Khuri F, et al. Lipid rafts and nonrafts mediate tumor necrosis factor–related apoptosis-inducing ligand–induced apoptotic and nonapoptotic signals in non–small cell Lung Carcinoma Cells. Cancer Res. 2007; 67: 6946-6955. Ref.: http://bit.ly/2J4J35m
- Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT-IIFADD-interacting protease. Fas/Apo-l. 1996; 803-815.
- Chinnaiyan AM1, Tepper CG, Seldin MF, O'Rourke K, Kischkel FC, et al. FADD/MORT1 is a common mediator of CD95 (Fas/APO-1) and tumor necrosis factor receptor-induced apoptosis. J Biol Chem. 1996; 271: 4961-4965. Ref.: http://bit.ly/2NuxCbk
- MacFarlane M. TRAIL-induced signalling and apoptosis. Toxicol Lett. 2003; 139: 89-97. Ref.: http://bit.ly/2Jnrmg3
- Degli-Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, et al. The novel receptor TRAIL-R4 induces NF-κB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity. 1997; 7: 813-820. Ref.: http://bit.ly/2LFh2mL
- Li H, Lin X. Positive and negative signaling components involved in TNFα-induced NF-κB activation. Cytokine. 2008; 41: 1-8. Ref.: http://bit.ly/308Su9l
- Trauzold A, Siegmund D, Schniewind B, Sipos B, Egberts J, et al. TRAIL promotes metastasis of human pancreatic ductal adenocarcinoma. Oncogene. 2006; 25: 7434. Ref.: http://bit.ly/2KSnGq4
- Ricci MS, Kim SH, Ogi K, Plastaras JP, Ling J, et al. Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell. 2007; 12: 66-80. Ref.: http://bit.ly/2J5frEL
- Secchiero P, Gonelli A, Carnevale E, Milani D, Pandolfi A, et al. TRAIL promotes the survival and proliferation of primary human vascular endothelial cells by activating the Akt and ERK pathways. Circulation. 2003; 107: 2250-2256. Ref.: http://bit.ly/2LyCkSD